Introduction
Use the Data Sources Preferences tab to
- view, add, or edit data source sites from which IGB can retrieve data.
- load personal genome version and chromosome synonyms files
- change how IGB caches data from remote and local sites
When you choose a species and genome version, IGB downloads meta-data about available genomes from each site listed the Data Sources list. If a site contains data for the currently displayed genome, then IGB displays those sites in the Available Data Sources/Data Sets section of the Data Access tab. And when you load sequence data into IGB by clicking the Load Sequence button, IGB attempts to obtain the sequence data from the data sources in the same order they appear in the Data Sources list.
To open the Data Sources Preferences tab
- Select Data Access > Configure
or
- Select File > Preferences > Data Sources
Data source sub-sections explained
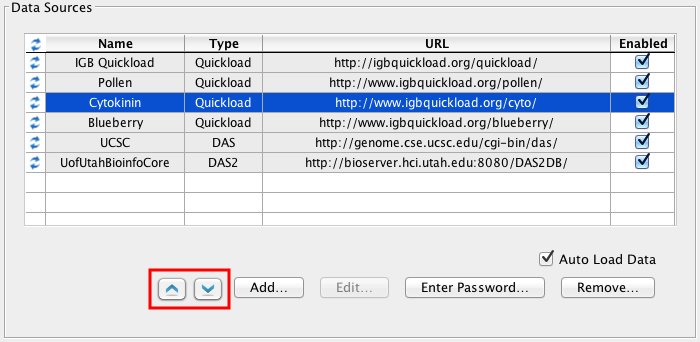
View, add, and configure data sources
This is the section where new data sources can be added, deleted or disabled. Extensive details about data source management can be found in Adding and Managing Data Source Servers.
Change sequence server order
Starting with IGB 6.6, you can change which data sources will provide sequence data. That is, if multiple data sources provide sequence data, the first one of the list will be used.
Click Up and Down buttons (red box) to change sequence server order.
Adding new data sources
Use Add.. to add a new data source.
Entering user names and passwords
Click Authentication.. to enter or change the user name and password IGB uses to access private data sources.
Most data sources don't require authentication. If you select a public (open) data source and click Authentication, IGB will allow you to enter a user name and password but won't use it when contacting the data source.
Change data loading settings
Some servers are configured to automatically load certain foundation data sets, such as the RefSeq gene models, into IGB as soon as the corresponding genome is selected. To block this behavior, uncheck Auto Load Data.
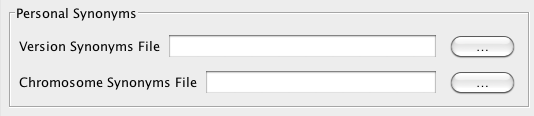
Personal synonyms files
Different data sources often refer to the same genome version or chromosome by different names.
For example, hg17, ensembl1834 and H_sapiens_May_2004 all refer to the same genome assembly and sequence. Similarly, some data providers use chr1 to refer to chromosome one while others may use 1 to mean the same thing.
IGB provides a system for designating synonyms for sequences and genome versions.
Genome synonyms File
A version synonyms file matches genome versions and allows you to display data from multiple sources in the same window.
Each time IGB restarts, synonyms will be loaded and merged from all the servers in use as well as from any personal synonyms file that you have specified.
A genome version synonyms file follows a simple tab-delimited format in which synonyms for the same genome appear in the same line, separated by tab characters.
Chromosome Synonyms File
A chromosome synonyms file is tab-delimited. Each line lists possible names for the same sequence.
Each time IGB starts, synonyms will be loaded and merged from all the servers in use as well as from any personal chromosome synonyms file entered here.
Cache Settings
IGB caches data that you have loaded over the web in order to speed the loading process the next time you use it. Files loaded from your local file system will not be cached.