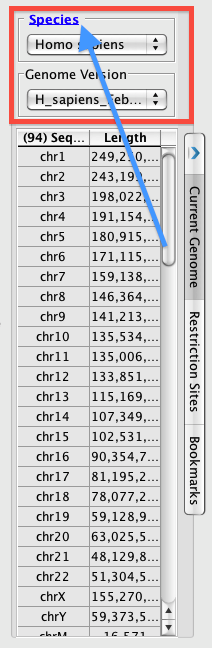
The Current Genome panel contains Species and Genome Version (red box). You can set the Current Genome panel to appear in either the left or right tray. The word 'Species' is now hyperlinked to return you to the starting page, with the picture selection (blue arrow). Otherwise, the drop boxes work as usual, where you pick your species, then you pick your genome version.
The sequences that are displayed in this tab can be sorted by name or by size. Clicking the column header Sequence(s) will sort by sequence name in alphabetical order. Clicking on Length will sort the sequences by size. An additional piece of information is the number next to Sequence(s), which indicates how many sequences are present in the genome version.
Most genome versions will have the option of being displayed as 'genome'. This option is found at the bottom of the Sequence(s) list, if it is available. Using this option displays every annotation in a linear arrangement, leaving a space between each chromosome/ contig (red arrows). In the coordinate axis, IGB will show you the relative length of each chromosome as black bars, with white space in between (red box). To display the entire genome, all of the data must be loaded. For organisms that have an autoloaded reference sequence, such as human, mouse, Drosophila, Arabidopsis, etc., you can select genome immediately (picture below shows the whole genome displayed for Arabidopsis). For other organisms, you will need to load each chromosome or contig individually.