Introduction
The Sequence Viewer gives users the option to view a region of sequence in a separate window with many options. The viewer will display the nucleotide sequence of selected sequence, exons, gene models, cytobands, or short reads. When you right-click a selected item in the display, such as a gene model, you'll see a menu offering View Genomic Sequence. Alternatively, you can access the same command from the View menu.
The viewer also has a variety of ways to display protein translations, and the ability to copy/export nucleotide sequence, annotated nucleotide sequence, protein sequence, etc.
View Sequence
By default, the viewer depicts bases in yellow on a black background. Use the Colors menu to change the color scheme to Blue on white (exons in blue and introns in black)
Selected Sequence or Cytobands
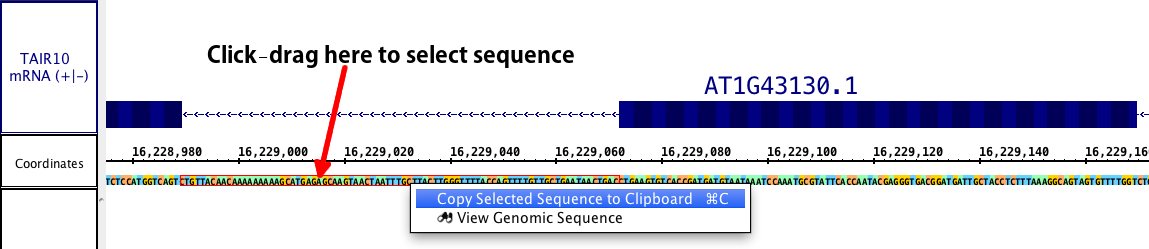
You can look at the selected sequence highlighted in the refseq, or (if present) the sequence of DNA contained in a cytoband. While this method loses the advantage of annotations/ exon designations, it does allow you to select DNA from introns, upstream and downstream regions, or any other area which may contain sequence of interest (i.e. promoters, enhancer binding elements, etc.). Select the sequence you wish to view by click-dragging the cursor over the sequence bar at any zoom level. A red outline will appear around the selected sequence.
This method of genomic DNA selection also allows a direct copy to the clipboard, if you wish to skip the Sequence viewer. Simply right click within the selected sequence, and chose Copy.
Right click within the selected area or open the View menu and chose View Genomic Sequence. The sequence viewer will open, and the title will show the genomic range of the sequence, the genome version, and the chromosome number. The Show cDNA option will be disabled, but all other options will remain. The DNA sequence will always be presented in the + (plus) direction. However, if your sequence of interest is the other strand, Show > Complement will provide the complement of the sequence (this is usable with all sequences).
The sequence viewer also allows you to save the reverse complement of the sequence you are viewing if you wish. Go to File > Save As FASTA (Reverse Complement).
Gene Models or Other Annotation
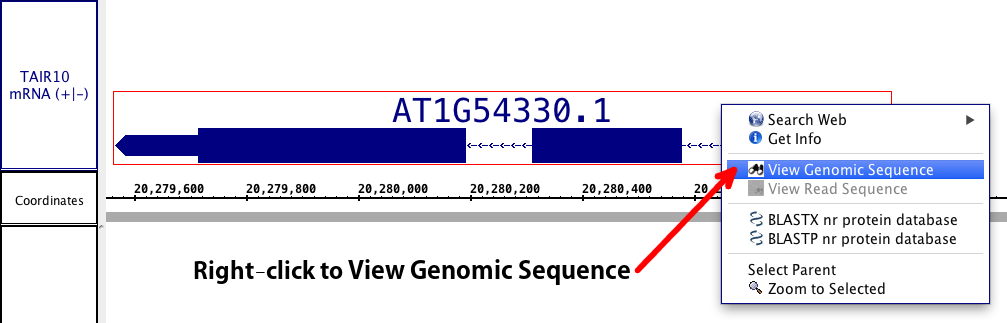
For gene models/ annotations, right click on the annotation or open View menu and choose option View Genomic Sequence.
To choose a single exon (part of) a gene model, right-click the exon. To choose the entire gene model, right-click any of the introns or the label, if available.
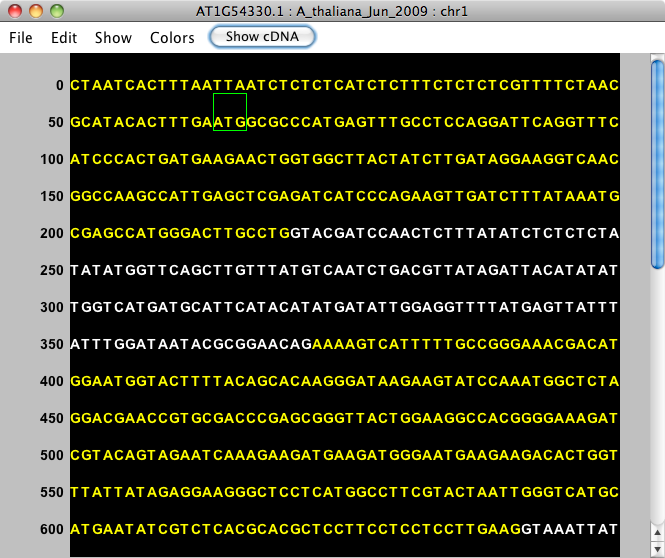
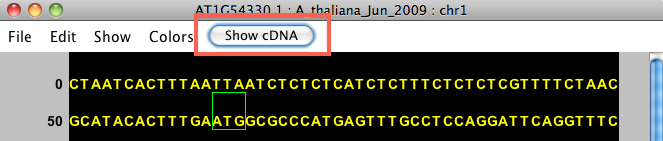
Choosing View Genomic Sequence opens the Sequence Viewer as shown below. By default, the viewer depicts exonic bases in yellow and intronic bases in white against a black background. The start codon (ATG) is shown with a green box around it, and the stop codon has a red box around it.
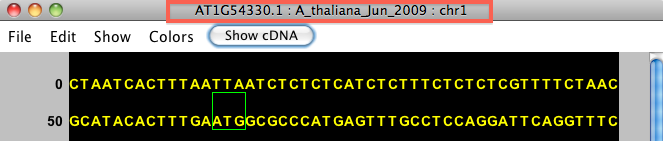
The title bar shows the name of the selected item. For instance, the image below shows genomic sequence for gene model AT1G54330.1, from the A_thaliana_Jun_2009 genome version, on chromosome 1.
Short Reads/ Alignments
The Sequence Viewer has a special option for short reads. If you select a short read and select View Genomic Sequence in Sequence Viewer, you will see the genomic sequence that the read is aligning to. The option View Read Sequence in Sequence Viewer will show the sequence of the read itself, including any inserted sequence. If a deletion is present, the deleted nucleotide(s) will appear as a dash (-).
Additional Features for Annotated Gene Models
Show cDNA
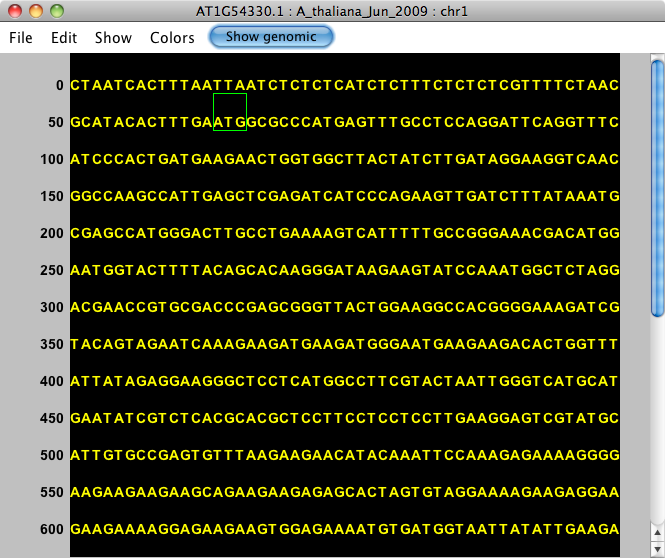
After selecting a gene model, the Sequence Viewer allows you to select Show cDNA, which will eliminate introns from the displayed sequence. We recommend selecting this option before using the translation feature. If an individual exon is selected, this button will be available, but will not change the sequence.
After selecting this feature, you'll see just the exonic (block) sequence, and the button will read Show genomic.
Show translation
Translation options are accessed from the Show menu. Reverse Complement provides the reverse complement in place (changes what is displayed). The Complement provides the complement under the main sequence. The + (forward) translations appears over the sequence; the - (reverse) translations below. To copy these translations, see Copy below
Copying and Saving Sequence
As mentioned, you can copy selected sequence to the clipboard directly from the main window of IGB. Additionally, you can copy a selection or all of your sequence from the sequence viewer. Highlight the sequence of interest (as shown below) and use Cop > Copy selected sequence to clipboard. Then open a text editor of choice and paste the sequence.
You can also Copy DNA with Lowercase Introns and Start/Stop Codons (NEW!) which copies the DNA sequence with the annotations. When you paste this sequence, you will be able to determine intron and exon boundaries, and fine the annotated start and stop codons within the nucleotide sequence. This can be very helpful for PCR primer design or alternative splicing studies.
Copy Annotated Translation (NEW!) will copy the annotated translation for the selected gene model - no highlighting needed.
Copy Selected Translation(s) (NEW!) requires that you Show 1-6 translations. The user then highlights the region of interest (possibly the whole sequence). This function then copies the translation(s). When pasted each translation is concatenated so that all of +1 shows together, then all of +2, then all of +3, etc. with a label indicating which translation is which.
You also have the option to save the sequence directly from the viewer into a fasta file. Fasta files can be shared and used for bioninformatics in this file format, but fasta files can also be opened and viewed from any text editor to access the sequence directly. The file will contain the sequence as well as a line with all of the relevant information from the title of the sequence viewer window. We allow you to save the sequence directly or in the reverse complement, as you need. Open File > Save as Fasta or Save as Fasta (reverse complement).
Finally, you can export an image of the viewer directly using the Save Image function.