Introduction
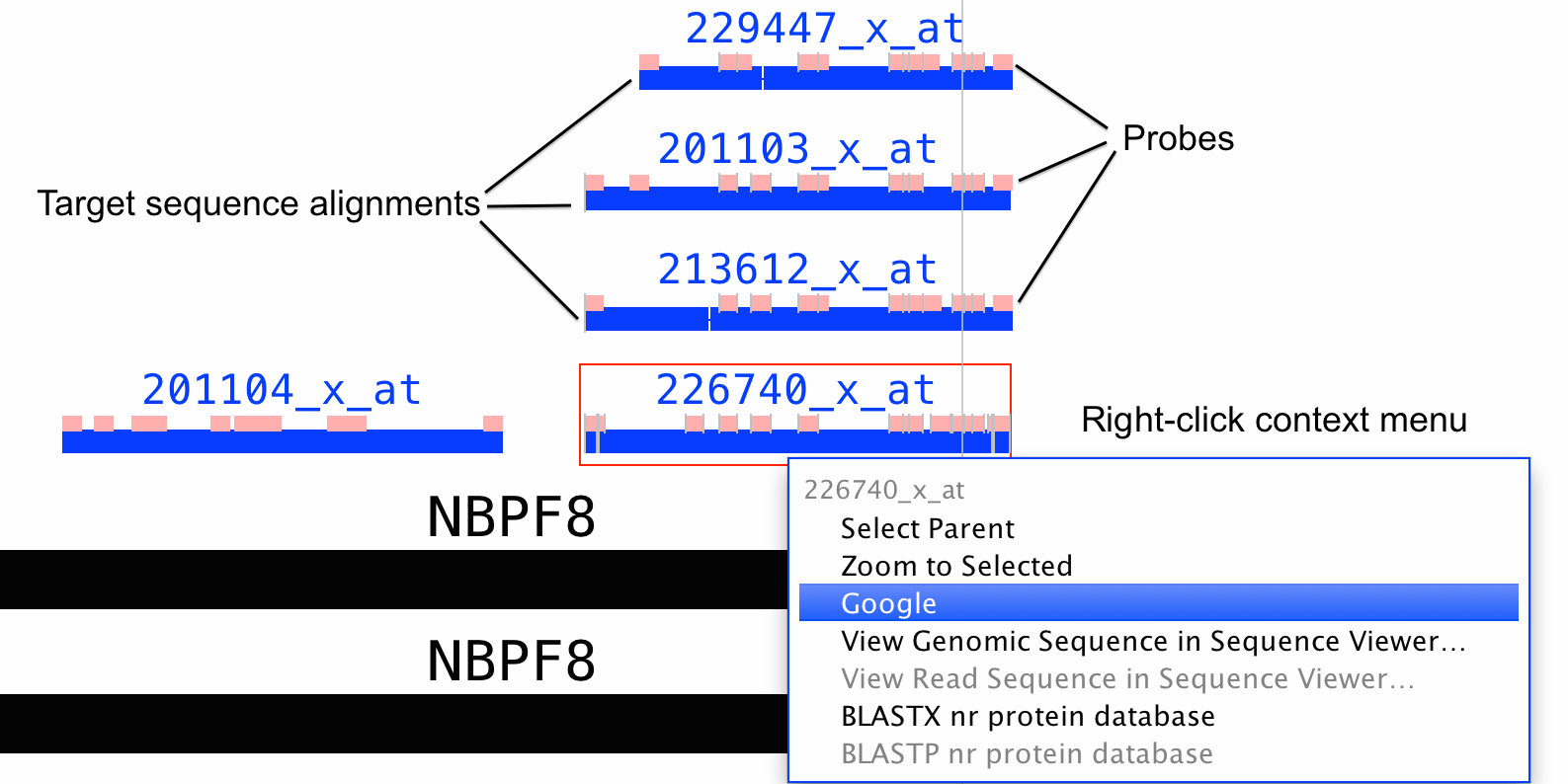
IGB can display Affymetrix probe sets aligned onto a reference genome - it can show probe set design sequences aligned onto a genome with the locations of the probes indicated as blocks.
When IGB was first developed at Affymetrix, the company distributed probe set alignment files for its catalog 3' IVT arrays. In recent years, however, they've stopped updating these files. So for some genomes, the alignment files you can find on the Affymetrix Web site reference obsolete reference genomes. If you need to work with more up-to-date genomes, we recommend you create your own alignment files or request them from the IGB team. For some genomes, we've added probe set alignments to the main IGBQuickLoad.org site. Mainly we've done this at the request of individual researchers, and so if you would like to request an array, let us know. If probe set alignments are available from our site, you'll typically find them in a folder named Affymetrix under the Data Sources section of the Data Access panel.
You can also make your own probe set alignment files using blat, tabix, and a python script we wrote. For more information, this Bitbucket repository.
About probe set alignment visualizations
Depending on when the arrays were designed, Affymetrix typically used expressed sequences from GenBank to select probes for probe sets. They then selected a region near the 3' end of each and then selected individual probes from that region. They distribute the sequences of probes in a probe set on their Web site, along with files labeled "target sequences" that contain the region from which the probes were selected. They also distribute what they call "exemplar" and "consensus" sequences taht contain the smaller target region from which the probes were selected.
Probe set visualization in IGB show the alignments of target, exemplar, or consensus sequences onto the genome. They also show the locations of probes that were selected from the design sequence. See the preceding figure for an example.
Because probes were selected from the expressed sequences, sometimes a probe will be shown as split across an intron. Also, sometimes probes overlap. And sometimes probes may be missing. If the target sequence contains a region that can't align onto the reference, and if this unaligned sequence contains a probe, then that probe will not be shown.
If you have questions about what you see in a probe set alignment, let us know.