Introduction
This tutorial describes how to import sequence and annotations into IGB when your genome of interest is not available from an IGB QuickLoad or Distributed Annotation site.
In this tutorial, we will demonstrate importing a bacterial genome (E. coli) using files downloaded from NCBI. Overall, to view custom genome and annotations in IGB properly a Synonym File needs to be created.
Get genome data from NCBI
First visit NCBI to retrieve the sequence data and annotations.
- Go to the NCBI GenBank record for E. coli K-12 subtr. MG1655: http://www.ncbi.nlm.nih.gov/nuccore/NC_000913.3
Download annotations file
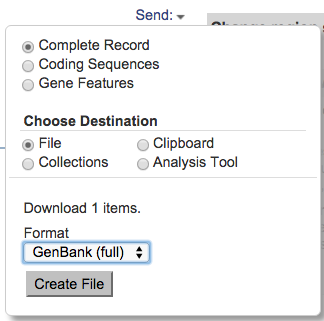
- Click Send menu (on the right side of the record)
- Select Complete Record, File, GenBank(Full), and Create File
Save the file to your computer; by default, it will be named "sequence.gb".
If you change the name, be sure to use the ".gb" file extension so that IGB can recognize the file and the file format.
Download sequence file
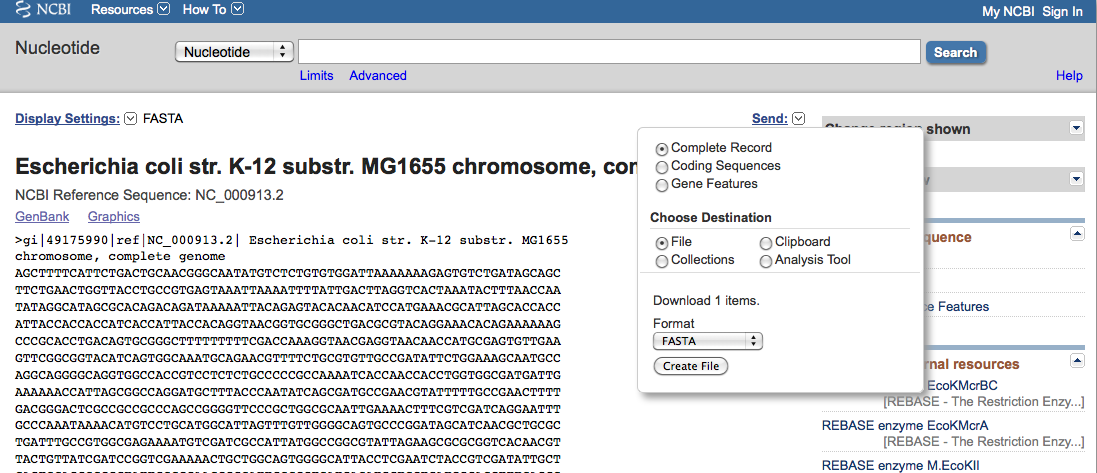
- Click on the FASTA link (on the left side of the record)
- Click Send menu
- Select, Complete Record, File, FASTA, and Create File
Save the file to your computer; by default, it will be named "sequence.fasta".
Make sure that the extensions for the FASTA file is either '.fasta' or '.fa'. If the extension is '.txt', you can safely change it to '.fa'.
Create Synonyms File
When IGB shows the sequence.gb file, it uses the 'LOCUS' name from the sequence.gb. To show the sequence.fasta file, IGB uses the 'sequence name' that follows the >. Note, when the labels for each file type (FASTA sequence files, annotation files, .bam/alignment files, etc.) for the exact same chromosome/sequence are all different IGB will treat each one as a separate chromosome; they will not be visualized together.
To overcome this, we enter 'synonyms'; IGB already has some internal synonyms, for example "1" and "chr1" are equivalent. You will need to know the sequence names of each of these files; if you are not sure, a quick way is to drag all of the files into IGB at the same time.
Determine 'sequence names' in sequence and annotation files:
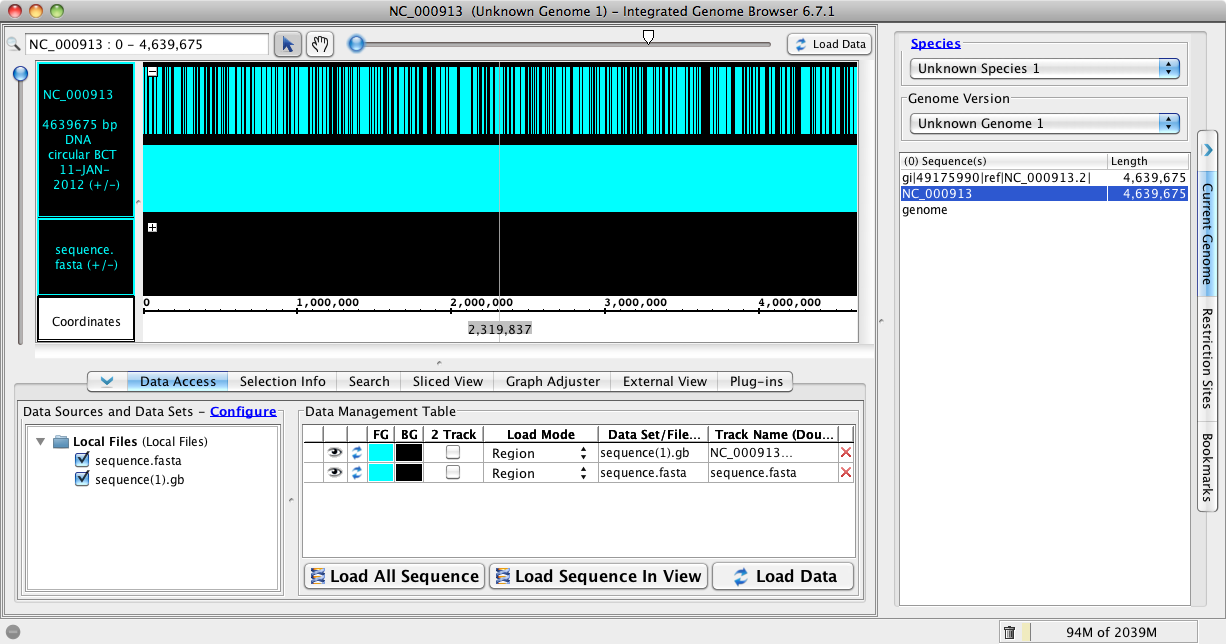
- Drag into IGB the sequence.fasta and sequence.gb
- In Current Genome Panel look under Sequences
- Note the sequences.fasta is called gi|556503834|ref|NC_000913.3| and the sequence.gb is labelled NC_000913
Create synonym file
Now that we know the headers of the files we will create a tab-delimited 'personal synonym' file:
- Open a Text Editor
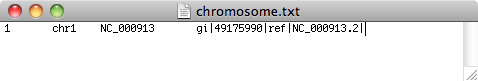
- On the first line, type 1 then press tab. Type chr1 then press tab
- 1 and chr1 are standard names for the first chromosome, and many files use these as their headers. Typically, we always include these two options in a synonym file
- Type in the header of the sequence.gb file,NC_000913, then press tab
- Type in the header of the sequence.fasta file, gi|556503834|ref|NC_000913.3|
- Save this file with the name chromsome.txt
If you are making a synonym file for a multi-chromosomal organism, then make a new line in the file for each chromosome, and just add all of the 'names' associated with it (make sure that there is a 'tab' between each name!). If you include a file that has a new name, open the chromosome.txt, and add the name to the proper line
Import Synonym File into IGB
In IGB:
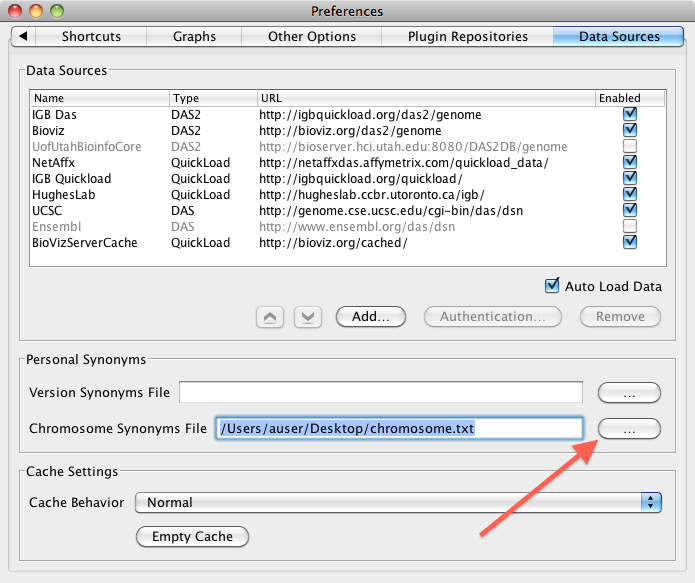
- Select Configure (under Data Access panel) to open the Preferences Menu
- Select Data Sources Tab
- Next to Chromosome Synonyms File browse for the chromosome.txt file
- Close the Preferences Menu
- Restart IGB
Visualizing the Reference Sequence and Models
When you open the new instance of IGB, your synonyms will be loaded for you. At this point, we will open the files, sequences.fasta and sequences.gb, so you can begin analysis of your own data.
To open the sequence and annotation files:
- File > Open Reference Sequence...
- Browse for the 'sequence.fasta' file.
- File > Open File...
- Browse for the 'sequence.gb' file.
- Click the Load Data button. NOTE: you need to click the Load Data button and NOT the Load Sequence button to visualize your sequence file!
Both the sequence and the models will load. You will be zoomed out, so the sequence will appear as a grey bar; as you zoom in the colors and nucleotides will become visible. At this point, you can begin viewing the gene models and sequence. Keep in mind that if the name of the chromosome(s) is different in your files, you will have to add the name to the chromosome.txt and then reopen IGB so it can load in the new information.